13.10.1 Last Universal Ancestor
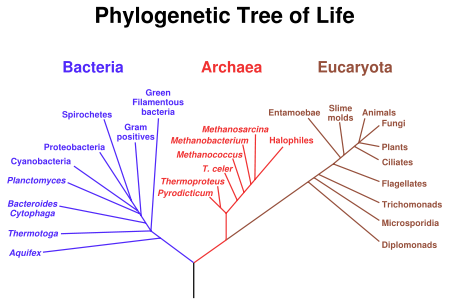
A cladogram linking all major groups of living organisms to the LUA (the black trunk at the bottom). This graph is derived from ribosomal RNA sequence data.
The last universal ancestor (LUA, also called the last universal common ancestor, LUCA, the cenancestor or “number one” in slang) is the most recent organism from which all organisms now living on Earth descend. Thus it is the most recent common ancestor (MRCA) of all current life on Earth. The LUA is estimated to have lived some 3.5 to 3.8 billion years ago (sometime in the Paleoarchean era).
13.10.2 Features
Based on the properties shared by all independently living organisms on Earth:
- The genetic code is based on DNA.
- The DNA is composed of four nucleotides (deoxyadenosine, deoxycytidine, deoxythymidine and deoxyguanosine), to the exclusion of other possible deoxynucleotides.
- The genetic code is composed of three-nucleotide codons, thus producing 64 different codons. Since only 20 amino acids are used, multiple codons code for the same amino acids. This structure is arbitrary and shared by all eukaryotes and prokaryotes. Archaea and mitochondria use a similar code with minor differences.
- The DNA is kept double-stranded by a template-dependent DNA polymerase.
- The integrity of the DNA is maintained by a group of maintenance enzymes, including DNA topoisomerase, DNA ligase and other DNA repair enzymes. The DNA is also protected by DNA-binding proteins like histones.
- The genetic code is expressed via RNAintermediates, which are single-stranded.
- RNA is produced by a DNA-dependent RNA polymerase using nucleotides similar to those of DNA with the exception of Thymidine in DNA, replaced by Uridine in RNA.
- The genetic code is expressed into proteins. All other properties of the organism (e.g. synthesis of lipids or carbohydrates) are the result of protein enzymes.
- Proteins are assembled from free amino acids by translation of an mRNA by ribosomes, tRNAand a group of related proteins.
- Ribosomes are composed of two subunits, one big and one small.
- Each ribosomal subunit is composed of a core of ribosomal RNA surrounded by ribosomal proteins.
- The RNA molecules (rRNA and tRNA) play an important role in the catalytic activity of the ribosomes
- Only 20 amino acids are used, to the exclusion of countless non-standard amino acids; only the L-isomersare used.
- Amino acids must be synthesized from glucose by a group of specialized enzymes; the synthesis pathways are arbitrary and conserved.
- Glucosecan be used as a source of energy and carbon; only the D-isomeris used.
- Glycolysis goes through an arbitrary degradation pathway.
- ATP is used as an energy intermediate.
- The cell is surrounded by a cellular membrane composed of a lipid bi-layer.
- Inside the cell, the concentration of sodium is lower, and potassium is higher, than outside. This choice is arbitrary. This gradient is maintained by specific ion pumps.
- The cell multiplies by duplicating all its contents followed by cellular division.
13.10.3 Lowest Common Ancestor
The lowest common ancestor (LCA) is a concept in graph theory and computer science. Let T be a rooted tree with n nodes. The lowest common ancestor is defined between two nodes v and w as the lowest node in T that has both v and w as descendants (where we allow a node to be a descendant of itself).
The LCA of v and w in T is the shared ancestor of v and w that is located farthest from the root. Computation of lowest common ancestors may be useful, for instance, as part of a procedure for determining the distance between pairs of nodes in a tree: the distance from v to w can be computed as the distance from the root to v, plus the distance from the root to w, minus twice the distance from the root to their lowest common ancestor.
In a tree data structure where each node points to its parent, the lowest common ancestor can be easily determined by finding the first intersection of the paths from v and w to the root. In general, the computational time required for this algorithm is O(h) where h is the height of the tree (length of longest path from a leaf to the root). However, there exist several algorithms for processing trees so that lowest common ancestors may be found more quickly, in constant time per query after a linear time pre-processing stage.
13.10.4 History
Dov Harel and Robert Tarjan (1984) were the first to develop an efficient lowest common ancestor data structure. Their algorithm processes any tree in linear time, so that subsequent lowest common ancestor queries may be answered in constant time per query. Tarjan also found a simpler but less efficient algorithm, based on the union-find data structure, for computing lowest common ancestors of an offline batch of pairs of nodes.
Baruch Schieber and Uzi Vishkin (1988) simplified the data structure of Harel and Tarjan. Their simplification is based on the principle that, in two special kinds of trees, lowest common ancestors are easy to determine:
– If the tree is a path, then the lowest common ancestor can be computed simply from the minimum of the levels of the two queried nodes.
– If the tree is a complete binary tree, the nodes may be indexed in such a way that lowest common ancestors reduce to simple binary operations on the indices.
The structure of Schieber and Vishkin decomposes any tree into a collection of paths, such that the connections between the paths have the structure of a binary tree, and combines both of these two simpler indexing techniques.
Omer Berkman and Uzi Vishkin (1993) discovered a completely new way to answer lowest common ancestor queries, again achieving linear pre-processing time with constant query time. This method was later rediscovered and simplified by Michael Bender and Martin Farach-Colton (2000). As had been previously observed by Gabow, Bentley & Tarjan (1984), the range minimum problem can in turn be transformed back into a lowest common ancestor problem using the technique of Cartesian trees.
13.10.5 Evolutionary developmental biology
Evolutionary developmental biology is a field of biology that compares the developmental processes of different animals and plants in an attempt to determine the ancestral relationship between organisms and how developmental processes evolved. It addresses the origin and evolution of embryonic development and shows:
– How modifications of development and developmental processes lead to the production of novel features
– The role of developmental plasticity in evolution
– How ecology impacts in development and evolutionary change
– The developmental basis of homoplasy and homology.
The contemporary field of evo-devo has gained impetus from the discovery of genes regulating embryonic development in model organisms. General hypotheses remain hard to test because organisms differ so much in shape and form. Nevertheless, it now appears that just as evolution tends to create new genes from parts of old genes, evo-devo demonstrates that evolution alters developmental processes to create new and novel structures from the old gene networks or will conserve a similar program in a host of organisms. Initially the major interest has been in the evidence of homology in the cellular and molecular mechanisms that regulate body plan and organ development. However more modern approaches include developmental changes associated with speciation.

Be First to Comment